Signaling crosstalk in the colon cancer microenvironment
Our research group is investigating the cellular and molecular processes in the development of colorectal cancer. In particular, we are interested in the communication of different cell
types in the immediate surrounding of the tumor, the so-called “tumor microenvironment”. For this purpose, we use “organoids”, a novel three-dimensional tissue culture system.
Experimental models for the human CRC microenvironment
Patient-derived tumor organoids and co-culture models
Colorectal cancer (CRC) is a leading cause of cancer-related deaths and is characterized by multiple genetic and epigenetic alterations. The individual tumor phenotype is strongly influenced by the tumor microenvironment that includes surrounding fibroblasts, vasculature, and immune cells. To study this complex disease and develop personalized therapeutic strategies a comprehensive approach is required.
Patient derived organoids (PDOs) have emerged as a research tool that mimics the 3D organization of the gastrointestinal epithelium. Furthermore, co-cultures with stromal cells can be performed to recreate a tissue-like context. PDOs can be expanded and cryopreserved as “living biobanks” that represent the tumor heterogeneity among patients. In collaboration with the University Tumor Center Frankfurt and supported by Frankfurt Cancer Institute (FCI), we are operating the FCI Organoid Biobank to provide support for culture and analysis of PDOs.
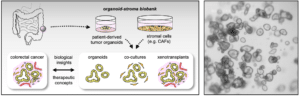
Genetic regulators of the CRC microenvironment
Tissue homeostasis depends on the interaction of stem cells with signals from the microenvironment. In tumors, this interaction is perturbed causing uncontrolled growth. To study tissue crosstalk we employ organoid co-cultures and transplantation models. Recently, we have identified that tumor mutations that activate Wnt signaling also influence cancer-associated fibroblasts (CAFs). Wnt inhibition induces a CAF subtype that renders tumor cells more invasive (Mosa et al., Cancer Research 2020), highlighting how stromal cell plasticity can regulate tumor malignancy.
To study influences of the tumor microenvironment on a global level we have performed genetic screens in human colon organoids using the CRISPR/Cas9 technology. This allows unbiased identification of tumor suppressor genes that promote growth in vitro and after organoid xenotransplantation (Michels et al., Cell Stem Cell 2020). In future, this platform could help to discover individualized therapeutic targets for colon cancer.
Personalized models for CRC therapy and pharmacogenomic screening
PDOs can serve as a preclinical model to identify therapeutic strategies. In collaboration with Prof. Wels (Georg-Speyer-Haus), we have used chimeric antigen receptors (CAR)-modified NK cells to induce cytotoxicity of CRC cells. We have developed enzymatic and live imaging assays to evaluate efficacy CAR cells (Schnalzger et al., EMBO Journal 2019).
As participant of the EU-consortium “EUbOPEN” (Enabling and unlocking biology in the OPEN, 2020-2024), we have developed a PDO drug screening platform. The EUbOPEN consortium is funded by the Innovative Medicines Initiative (IMI2) and aims to generate an open access chemogenomic library of compounds covering the “druggable human genome”. Together with our partners from academia and pharmacologic industry, we develop “Human
Tissue Assays” for CRC. We conduct high-throughput pharmacologic screens using the organoid biobank to identify resistance mechanisms and therapeutic strategies.
The EUbOPEN project has received funding from the Innovative Medicines Initiative 2 Joint Undertaking under grant agreement No 875510. This Joint Undertaking receives support from the European Union’s Horizon 2020 research and innovation programme, EFPIA companies and Associated Partners: KTH, OICR, Diamond and McGill. This communication reflects the views of the authors and neither IMI nor the European Union, EFPIA or any Associated Partners are liable for any use that may be made of the information contained herein.